Clinical Annotations for Automatic Stuttering Severity Assessment

Our research addresses this critical gap by enhancing the FluencyBank dataset with comprehensive clinical annotations created by expert speech-language pathologists. Rather than relying on simplified or non-expert labeling, we prioritized clinical validity by hiring certified clinicians to provide multi-modal annotations that capture the full complexity of stuttering assessment. This work represents a significant step toward building machine learning models that can genuinely support clinical practice and improve accessibility to quality stuttering assessment.
The enhanced dataset includes detailed annotations for stuttering moments, secondary behaviors, and tension scores using established clinical standards. By incorporating both audio and visual features, our annotations provide a comprehensive foundation for developing robust automatic assessment tools. Most importantly, we provide expert consensus annotations for reliable model evaluation, ensuring that automated systems can be properly validated against clinical expertise.
Enhanced Clinical Annotations and Dataset
| Assessment Component | Clinical Focus | Annotation Details |
|---|---|---|
| Stuttering Moments | Core disfluencies (repetitions, blocks, prolongations) | Precise temporal boundaries and type classification |
| Secondary Behaviors | Associated physical movements | Multimodal detection using audiovisual features |
| Tension Scores | Severity rating of physical struggle | Clinical rating scales applied by expert annotators |
| Consensus Labels | Highly reliable ground truth | Multiple expert agreement for evaluation standards |
Inter-Annotator Agreement and Reliability
To ensure our annotations are reliable, we systematically calculated inter‑annotator agreement across temporal spans, disfluency types, and tension scores. Here’s how we approached it:
1. Grouping / Aligning Annotations
- We applied agglomerative clustering on annotated time segments using Intersection-over-Union (IoU) as the distance measure.
- Annotations that overlapped sufficiently were clustered together, creating comparable units for agreement computation.
2. Applying Agreement Metrics
For each component, we used Krippendorff’s α (alpha) — a robust reliability coefficient suitable for different data types and multiple annotators. it is computed as follows
Let Dₒ be the observed disagreement and Dₑ the expected disagreement:
For details, see Krippendorff’s explanation:
Krippendorff, K. (2013). “Computing Krippendorff’s Alpha-Reliability”.
The disagreements for each category are computed as follows:
| Component | Data Type | Distance/Similarity Metric | Krippendorff’s α |
|---|---|---|---|
| Temporal spans | Interval | 1 – IoU (interval distance) | 0.68 |
| Primary/Secondary types | Nominal | Binary distance (match/mismatch) | Varies by class |
| Tension scores | Ordinal | Normalized rank‑Euclidean distance | 0.18 |
You can compute α using our implementation on Github
Run the following commands
git clone https://github.com/rufaelfekadu/IAA.git
cd IAA
pip install numpy pandas matplotlib scikit-learnDownload the dataset from Here
wget https://raw.githubusercontent.com/mbzuai-nlp/CASA/refs/heads/main/data/Voices-AWS/total_dataset.csvUse the following script to compute the IAA
import pandas as pd
import numpy as np
import json
from agreement import InterAnnotatorAgreement
from granularity import SeqRange
input_path = "total_dataset.csv"
item_col = "item"
annotator_col = "annotator"
LABELS = ['SR','ISR','MUR','P','B', 'V', 'FG', 'HM', 'ME']
def binary_distance(x, y):
return 1 if x != y else 0
# euclidian distance
def euclidian_distance(a1, a2, max_value=3):
return abs(a1 - a2) / max_value
# Intersection Over Union
def iou(vrA, vrB):
xA = max(vrA.start_vector[0], vrB.start_vector[0])
xB = min(vrA.end_vector[0], vrB.end_vector[0])
interrange = max(0, xB - xA + 1)
unionrange = (vrA.end_vector[0] - vrA.start_vector[0] + 1) + (vrB.end_vector[0] - vrB.start_vector[0] + 1) - interrange
return (interrange / unionrange)
def inverse_iou(vrA, vrB):
return 1 - iou(vrA, vrB)
# Read the CSV file
grannodf = pd.read_csv(input_path)
results = {}
grannodf = grannodf[~grannodf['annotator'].isin(['Gold','bau','mas','sad'])]
print(grannodf['annotator'].unique())
grannodf['timevr'] = grannodf[['start','end']].apply(lambda row: SeqRange(row.to_list()), axis=1)
# compute IAA for each class
for label in LABELS[:-1]:
iaa = InterAnnotatorAgreement(grannodf,
item_colname=item_col,
uid_colname=annotator_col,
label_colname=label,
distance_fn=binary_distance)
iaa.setup()
results[label] = {
'alpha': iaa.get_krippendorff_alpha(),
'ks': iaa.get_ks(),
'sigma': iaa.get_sigma(use_kde=False),
}
# average number of annotaions per annotator for a given class
results[label]['num_annotations'] = grannodf[grannodf[label] == 1].groupby(annotator_col).size().mean()
# compute IAA for tension
iaa = InterAnnotatorAgreement(grannodf,
item_colname=item_col,
uid_colname=annotator_col,
label_colname='T',
distance_fn=euclidian_distance)
iaa.setup()
results['T'] = {
'alpha': iaa.get_krippendorff_alpha(),
'ks': iaa.get_ks(),
'sigma': iaa.get_sigma(use_kde=False),
'num_annotations': grannodf[grannodf['T'] >= 1].groupby(annotator_col).size().mean()
}
# compute IAA for time-intervals
iaa = InterAnnotatorAgreement(grannodf,
item_colname=item_col,
uid_colname=annotator_col,
label_colname='timevr',
distance_fn=inverse_iou)
iaa.setup()
results['time-intervals'] = {
'alpha': iaa.get_krippendorff_alpha(),
'ks': iaa.get_ks(),
'sigma': iaa.get_sigma(use_kde=False),
'num_annotations': grannodf.groupby(annotator_col).size().mean()
}
with open(f'iaa_results.json', 'w') as f:
json.dump(results, f, indent=4)3. Interpretation
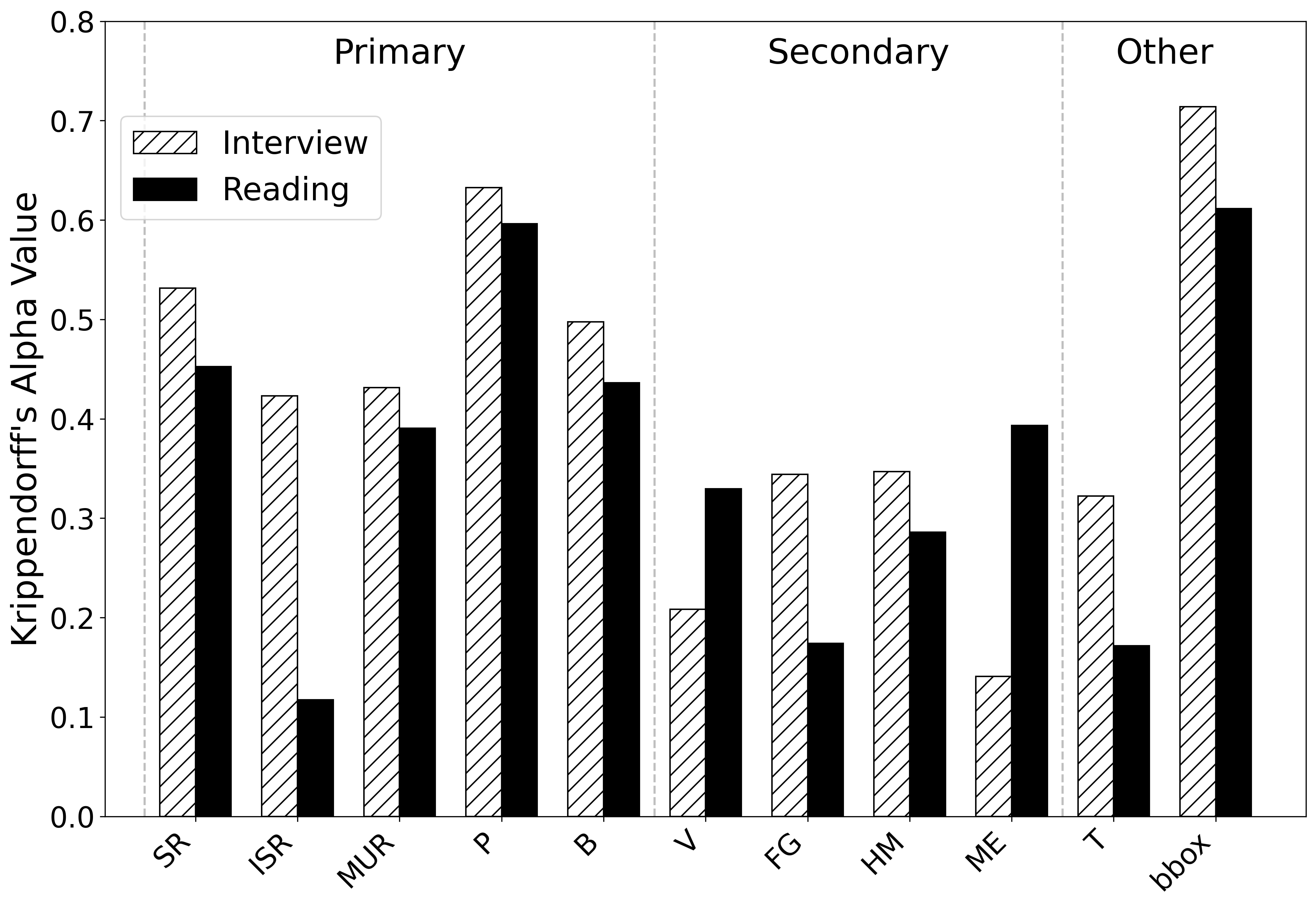
- A span α of 0.68 indicates substantial temporal agreement.
- Primary/secondary type α values vary by disfluency class, but generally show moderate consistency.
- The low tension score α (0.18) highlights the difficulty and subjectivity in rating physical struggle.
Applications and Expert Consensus Standards
Research Findings and Clinical Impact
Conclusion
The enhancement of FluencyBank with expert clinical annotations represents a significant advancement in stuttering research and assessment. By prioritizing clinical validity and expert knowledge, this work provides a foundation for developing automated assessment tools that can genuinely support clinical practice. The multi-modal annotation scheme, expert consensus process, and rigorous evaluation framework establish new standards for stuttering assessment research.
We encourage researchers and clinicians to explore the enhanced dataset and contribute to the ongoing development of clinically valid automatic stuttering assessment systems. Through continued collaboration between machine learning researchers and speech-language pathologists, we can work toward improving accessibility and quality of stuttering assessment and treatment services worldwide.
For access to the enhanced FluencyBank dataset and annotation guidelines, please visit our project repository and follow the clinical research protocols for responsible use of this sensitive clinical data.
Comments